pVACtools¶
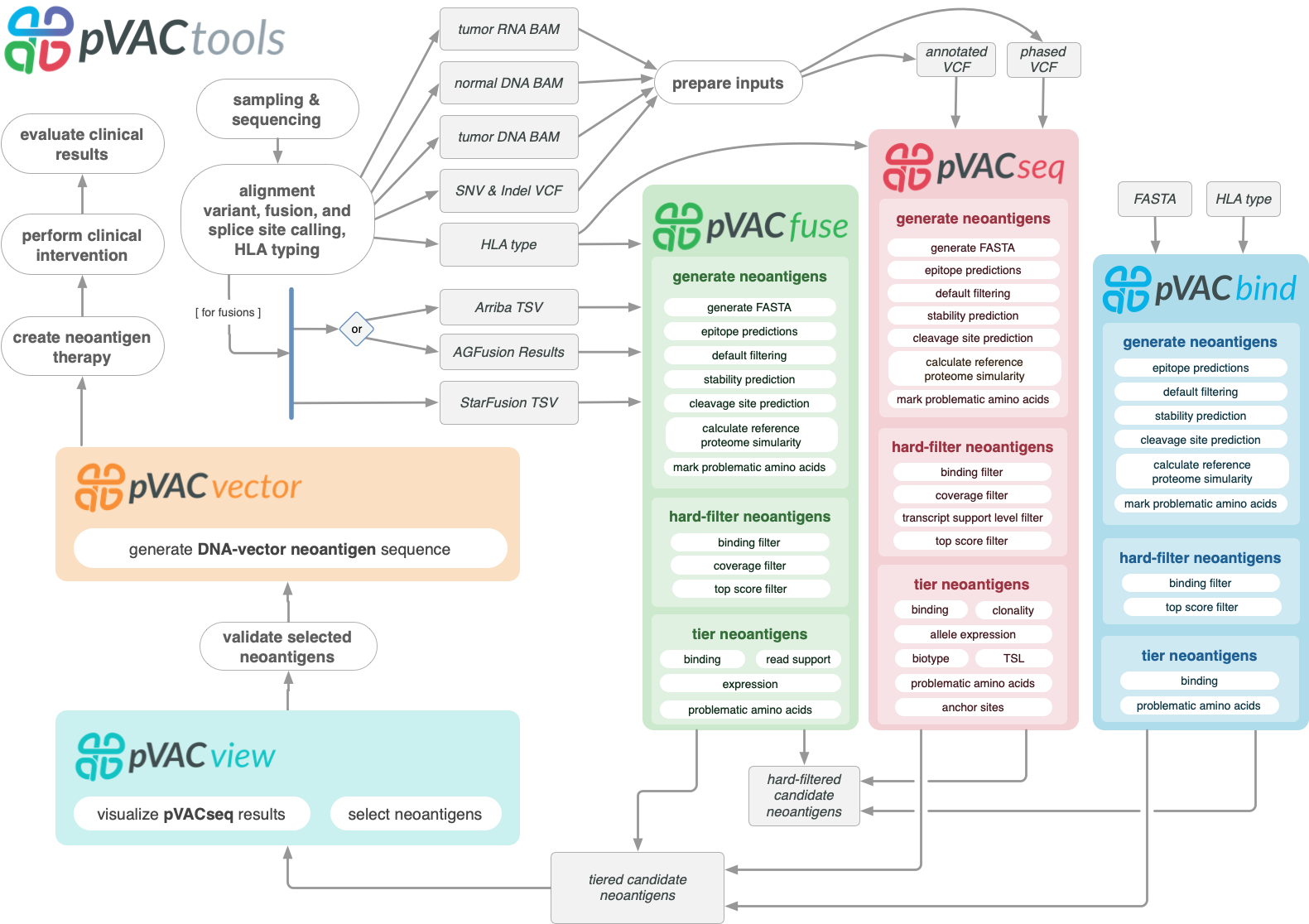
pVACtools is a cancer immunotherapy tools suite consisting of the following tools:
- pVACseq
A cancer immunotherapy pipeline for identifying and prioritizing neoantigens from a VCF file.
- pVACbind
A cancer immunotherapy pipeline for identifying and prioritizing neoantigens from a FASTA file.
- pVACfuse
A tool for detecting neoantigens resulting from gene fusions.
- pVACvector
A tool designed to aid specifically in the construction of DNA-based cancer vaccines.
- pVACview
An application based on R Shiny that assists users in reviewing, exploring and prioritizing neoantigens from the results of pVACtools processes for personalized cancer vaccine design.
Contents¶
New in Release 4.1.1¶
This is a bugfix release. It fixes the following problem(s):
The previous version updated how the all_epitopes.tsv file was parsed when creating the aggregated report and NA values are now parsed as native pandas nan. However, this update was not handled correctly for the Mutation Position column, leading to errors with NA values in that column. This release fixes this error. (#1079)
The previous version would write the DeepImmuno output file in the same location for multiple prediction calls. This would lead to errors when running in multi-threaded mode. This releases updates the code to write DeepImmuno outputs to unique file locations. (#1078)
This release updates how the list of combinatorial class II alleles is created in order to return it as a sorted list, creating a consistent order when writing the input.yml log file. (#1077)
This release updates the GitHub commit for the pVACview demo data in order to pull the latest version of this data, including DeepImmuno and BigMHC prediction data. (#1073)
This release fixes an issue where the pVACvector visualization images were saved in a low resolution format resulting in blurry images. (#1071)
This release fixes an issue where the method to determine the matched wildtype result didn’t return where appropriate, causing the mutation position to not be set correctly. (#1082)
This release fixes some typos. (#1072)
Past release notes can be found on our Release Notes page.
To stay up-to-date on the latest pVACtools releases please join our Mailing List.
Citations¶
Jasreet Hundal , Susanna Kiwala , Joshua McMichael, Chris Miller, Huiming Xia, Alex Wollam, Conner Liu, Sidi Zhao, Yang-Yang Feng, Aaron Graubert, Amber Wollam, Jonas Neichin, Megan Neveau, Jason Walker, William Gillanders, Elaine Mardis, Obi Griffith, Malachi Griffith. pVACtools: A Computational Toolkit to Identify and Visualize Cancer Neoantigens. Cancer Immunology Research. 2020 Mar;8(3):409-420. doi: 10.1158/2326-6066.CIR-19-0401. PMID: 31907209.
Jasreet Hundal, Susanna Kiwala, Yang-Yang Feng, Connor J. Liu, Ramaswamy Govindan, William C. Chapman, Ravindra Uppaluri, S. Joshua Swamidass, Obi L. Griffith, Elaine R. Mardis, and Malachi Griffith. Accounting for proximal variants improves neoantigen prediction. Nature Genetics. 2018, DOI: 10.1038/s41588-018-0283-9. PMID: 30510237.
Jasreet Hundal, Beatriz M. Carreno, Allegra A. Petti, Gerald P. Linette, Obi L. Griffith, Elaine R. Mardis, and Malachi Griffith. pVACseq: A genome-guided in silico approach to identifying tumor neoantigens. Genome Medicine. 2016, 8:11, DOI: 10.1186/s13073-016-0264-5. PMID: 26825632.
Source code¶
The pVACtools source code is available in GitHub.
License¶
This project is licensed under BSD 3-Clause Clear License.
