pVACtools¶
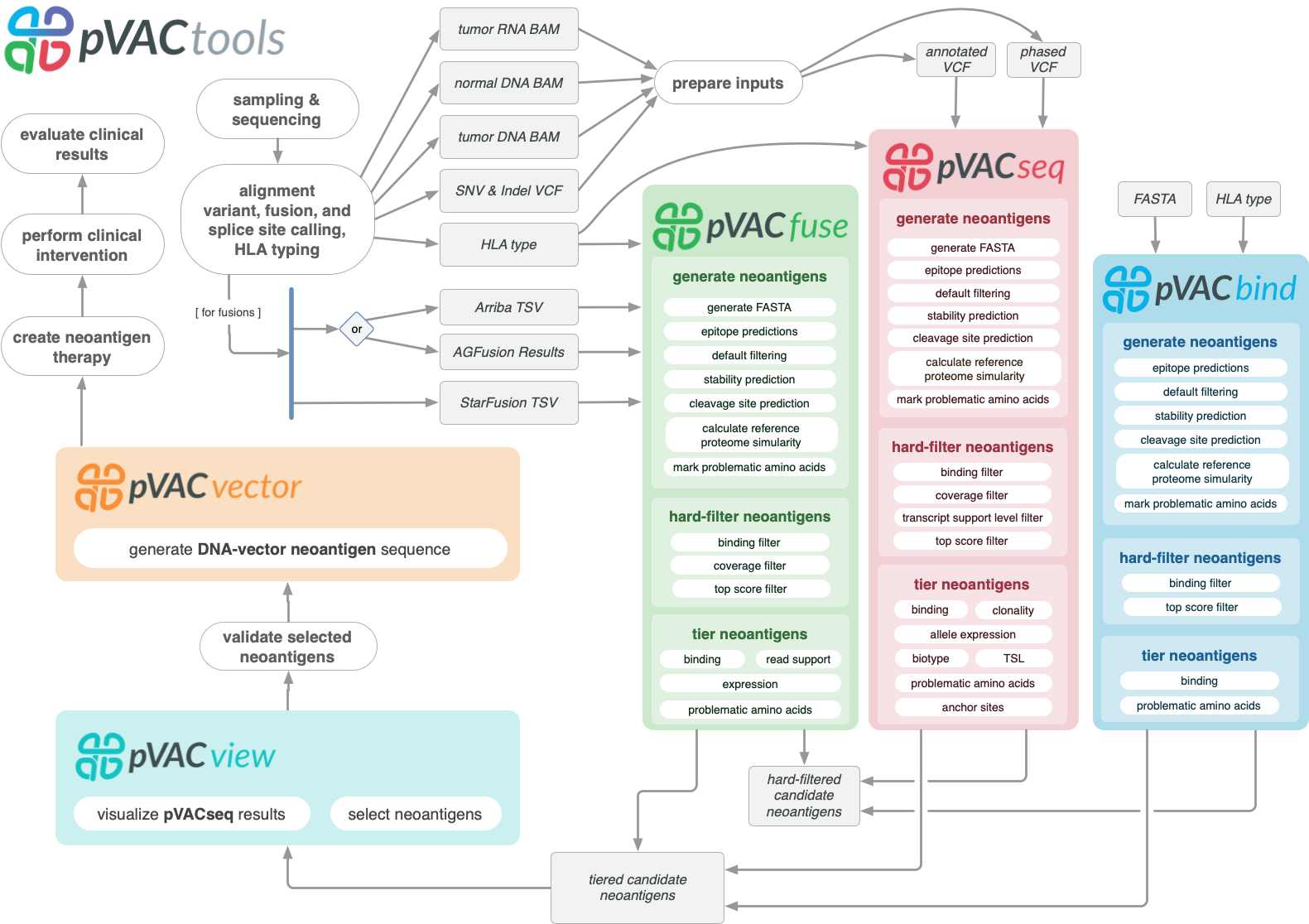
pVACtools is a cancer immunotherapy tools suite consisting of the following tools:
- pVACseq
A cancer immunotherapy pipeline for identifying and prioritizing neoantigens from a VCF file.
- pVACbind
A cancer immunotherapy pipeline for identifying and prioritizing neoantigens from a FASTA file.
- pVACfuse
A tool for detecting neoantigens resulting from gene fusions.
- pVACvector
A tool designed to aid specifically in the construction of DNA-based cancer vaccines.
- pVACview
An application based on R Shiny that assists users in reviewing, exploring and prioritizing neoantigens from the results of pVACtools processes for personalized cancer vaccine design.
Contents¶
New in Release 4.1.0¶
This is a minor feature release. It adds the following features:
This release adds support for two new prediction algorithms: BigMHC and DeepImmuno (#1063). BigMHC includes predictions for elution (BigMHC_EL) and immunogenicity (BigMHC_IM). DeepImmuno is a prediction algorithms for immunogenicty.
This release includes several updates to pVACview (#1012):
The tab containing the anchor heatmaps for each well-binding peptide of a variant has been moved to the “Transcript and Peptide Set Data” panel.
The anchor heatmap tab now also contains a table of all the per-length and per-allele anchor weights for each position in a peptide.
The pVACtools version is now displayed at the bottom of the sidebar.
A new panel has been added to show the tiering parameters currently applied.
Where there are multiple transcript sets for a variant, the one containing the best peptide is now selected by default.
This release adds a new command
pvactools download_wdls(#1055). This command can be used to download WDL workflow files for pVACseq and pVACfuse.
Past release notes can be found on our Release Notes page.
To stay up-to-date on the latest pVACtools releases please join our Mailing List.
Citations¶
Jasreet Hundal , Susanna Kiwala , Joshua McMichael, Chris Miller, Huiming Xia, Alex Wollam, Conner Liu, Sidi Zhao, Yang-Yang Feng, Aaron Graubert, Amber Wollam, Jonas Neichin, Megan Neveau, Jason Walker, William Gillanders, Elaine Mardis, Obi Griffith, Malachi Griffith. pVACtools: A Computational Toolkit to Identify and Visualize Cancer Neoantigens. Cancer Immunology Research. 2020 Mar;8(3):409-420. doi: 10.1158/2326-6066.CIR-19-0401. PMID: 31907209.
Jasreet Hundal, Susanna Kiwala, Yang-Yang Feng, Connor J. Liu, Ramaswamy Govindan, William C. Chapman, Ravindra Uppaluri, S. Joshua Swamidass, Obi L. Griffith, Elaine R. Mardis, and Malachi Griffith. Accounting for proximal variants improves neoantigen prediction. Nature Genetics. 2018, DOI: 10.1038/s41588-018-0283-9. PMID: 30510237.
Jasreet Hundal, Beatriz M. Carreno, Allegra A. Petti, Gerald P. Linette, Obi L. Griffith, Elaine R. Mardis, and Malachi Griffith. pVACseq: A genome-guided in silico approach to identifying tumor neoantigens. Genome Medicine. 2016, 8:11, DOI: 10.1186/s13073-016-0264-5. PMID: 26825632.
Source code¶
The pVACtools source code is available in GitHub.
License¶
This project is licensed under BSD 3-Clause Clear License.