
Output Files¶
File Name |
Description |
|---|---|
|
An intermediate file with vaccine peptide sequences created from the epitopes in a pVACseq output file. |
|
The final output file with the peptide sequences and best spacers in the optimal order. |
|
A JPEG visualization of the above result. |
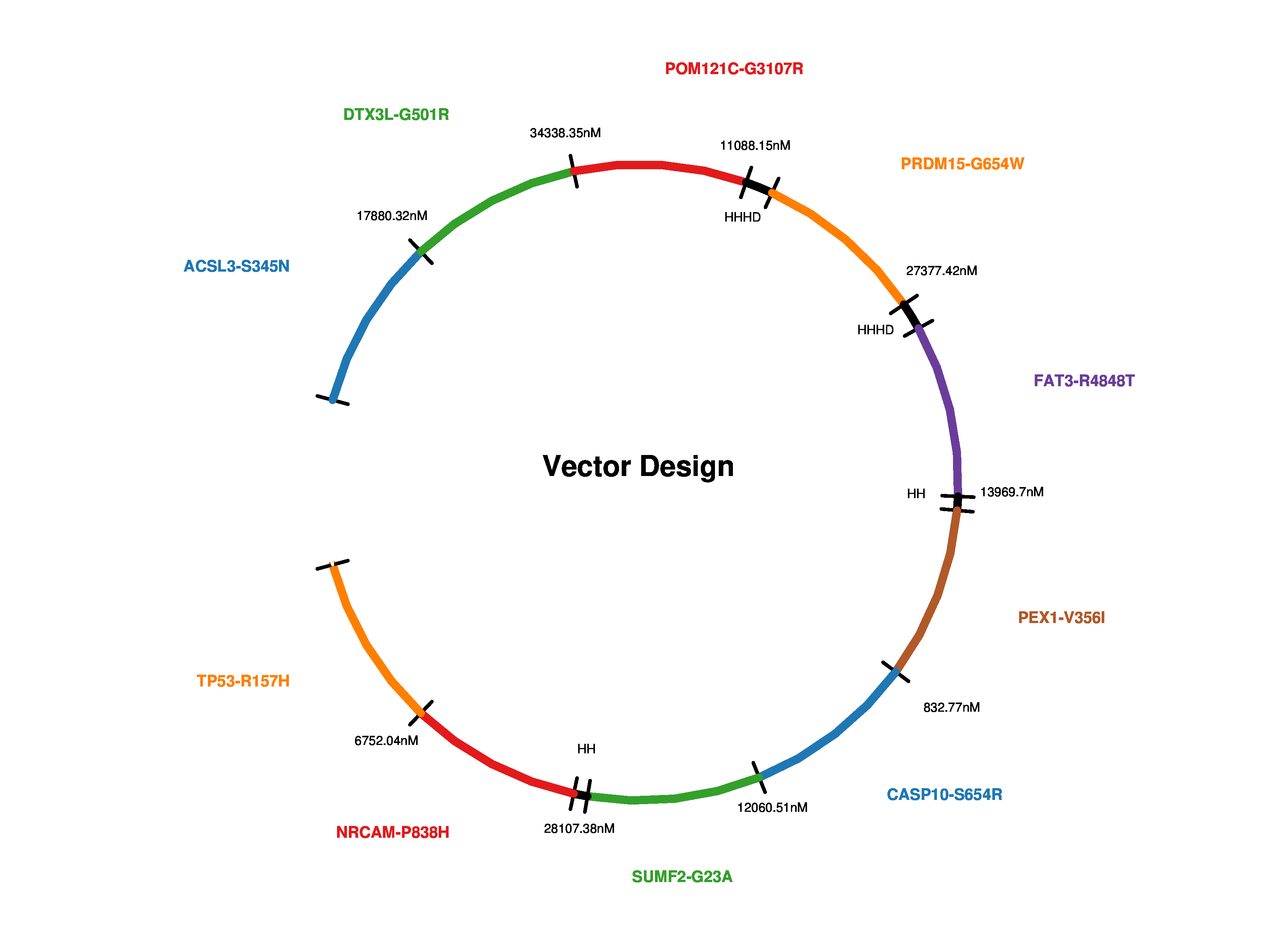
pVACvector result visualization example¶
